Enter a list of interactions. Every interaction in a separate line, as a pair of space-separated Uniprot Entries or Uniprot Accessions IDs
New Database with structural models of protein complexes derived by comparative modeling and data driven docking.
Enter a list of interactions. Every interaction in a separate line, as a pair of space-separated Uniprot Entries or Uniprot Accessions IDs
Enter a list of Uniprot Entries or Uniprot Accessions.
Users can retrieve the information of protein complexe querying by a particular pairs of proteins using the Uniprot entry gene ID space separated:
ARHGB_HUMAN RHOA_HUMAN
NXT2_HUMAN UBP54_HUMAN
Besides using a given pair(s), users can retrieve all the interactions for a given protein providing also the Uniprot entry gene ID:
NXT2_HUMAN
AKTIP_HUMAN
The server returns a list of structural models that users can browse including the visualization of the structure of the complex on a Mol* applet.
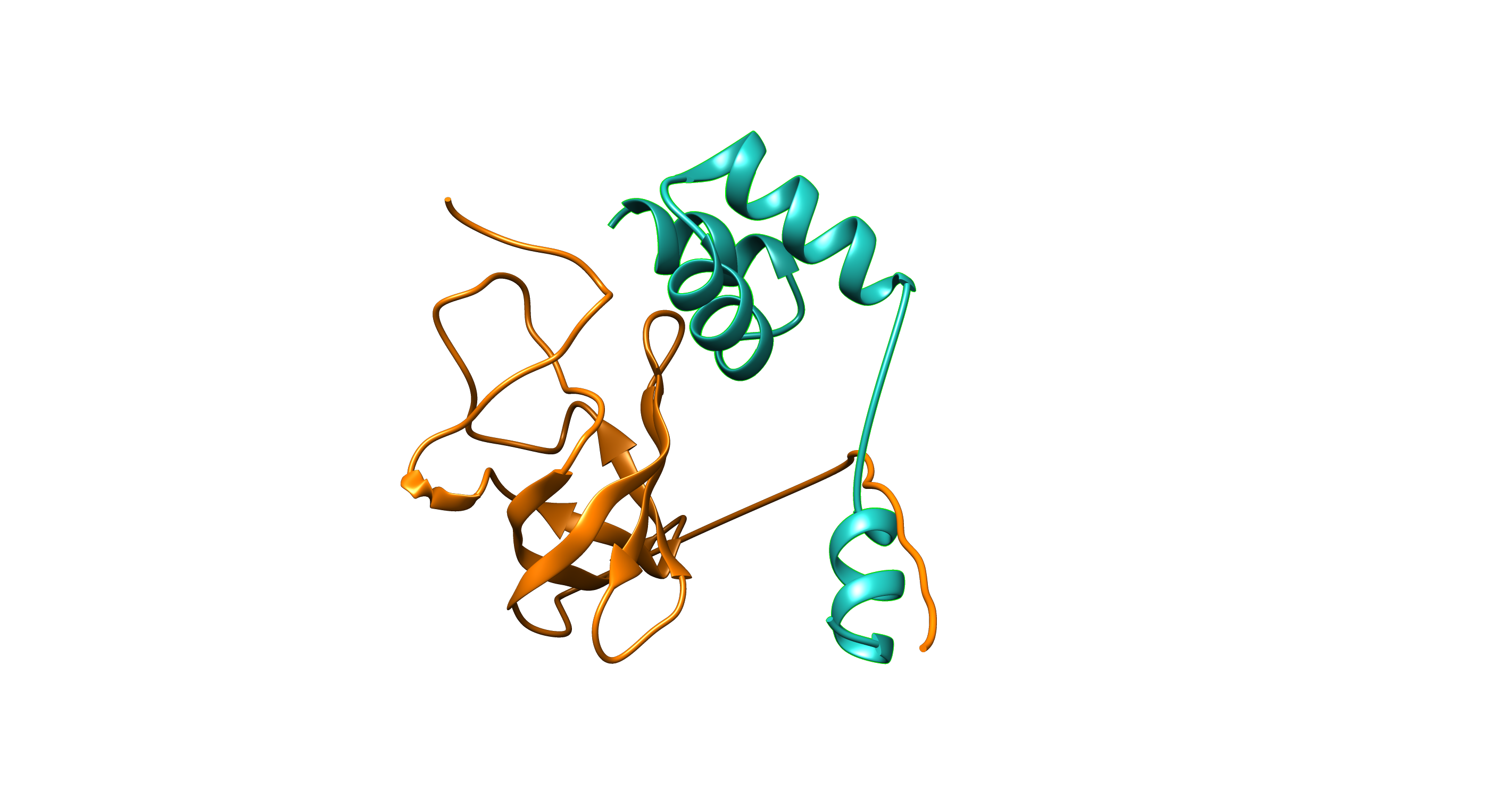
Interactome CM2D3 (Comparative Modeling and Data-Driven Docking) database.
This resource compiles structural models of protein complexes derived both by comparative modeling and docking using our original methods MODPIN and VD2OCK.

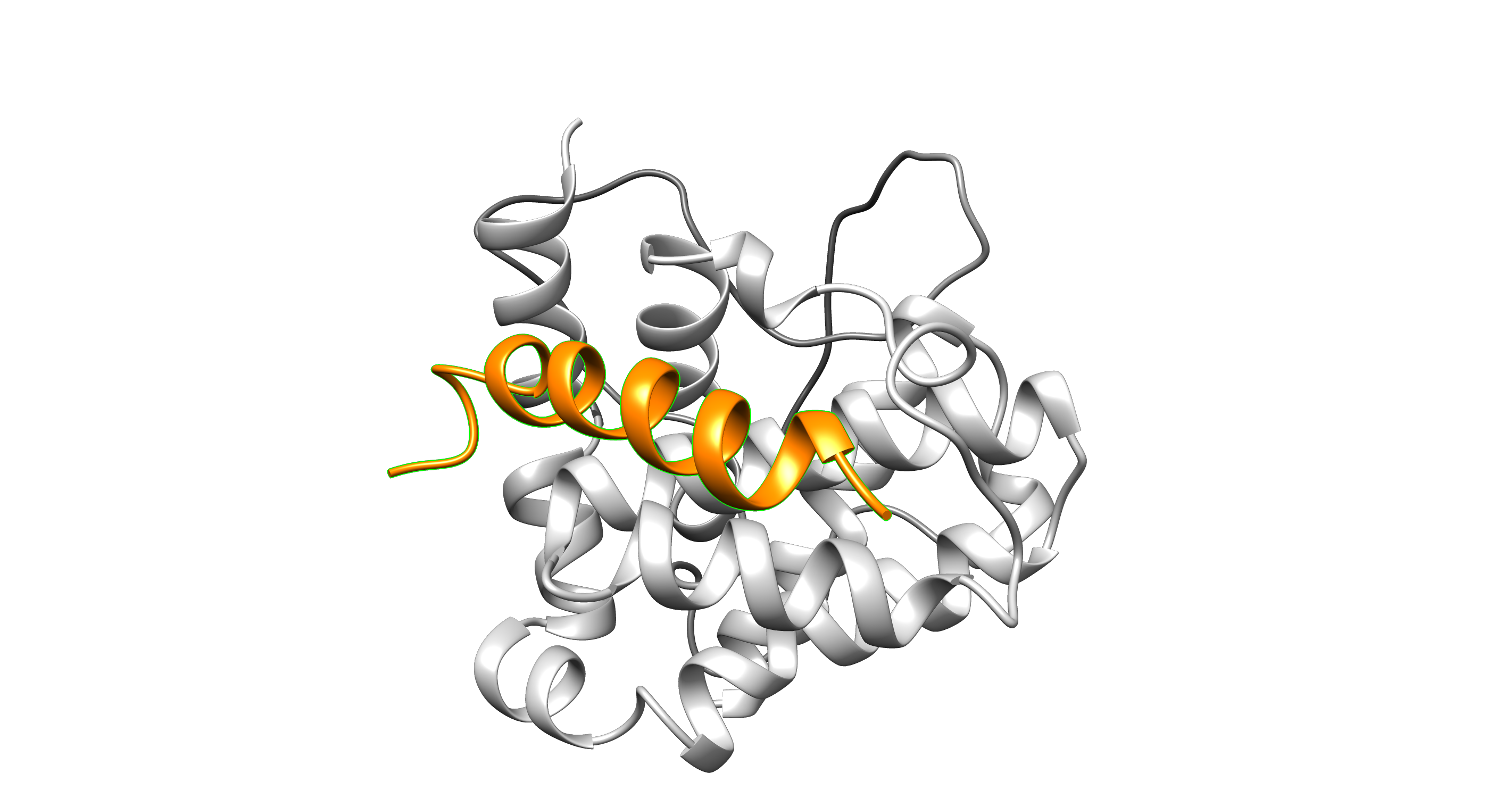
Homology modeling with MODPIN, a set of python script to model and analyze protein-protein interactions.
Protein docking with VD2OCK, a guided protein-docking algorithm to model quaternary structure of binary complexes.